
Innovative Analytical Ultracentrifugation Techniques for the Characterization of AAV Vectors
The therapeutic use of adeno-associated virus (AAV) accounts for the largest share of the global gene therapy industry due to its safety profile and proven efficacy in treating genetic diseases. Over the past 30 years, scientists, clinicians, and biotech industry professionals have worked with contract development and manufacturing organizations (CDMOs) to harness the natural abilities of AAV to deliver genetic information to specified cell types. The collaboration between AAV gene therapy developers and CDMOs has resulted in scalable manufacturing solutions to transition “a gene and a dream” into a biological therapeutic poised for successful patient outcomes and a greater number of commercialized treatments.
Generation, Characterization, and Removal of Empty AAV Capsids
Manufacturing processes are continuously optimized to ensure efficient, safe, and pure AAV gene therapy products are consistently delivered to patients. However, within a production cell line, AAV assembly and DNA packaging processes naturally lead to a mixture of full, empty, and some partial genome-containing capsid populations. While it has been demonstrated that AAV containing partial genomes can retain a level of therapeutic benefit, it is often with reduced potency in comparison to full genome containing capsids1. Additionally, empty AAV particles contain no therapeutic benefit while increasing the overall viral load and risk of immune response. Furthermore, a heterogeneous population of genome fragments within a final AAV drug product can be expected to show inconsistencies from lot-to -lot and cause issues when attempting to define critical quality attributes of the product. For this reason, capsids lacking a full vector genome are considered process impurities and are reduced through various methods during downstream processing.
Optimizing full capsid enrichment techniques, which may lead to achieving higher therapeutic efficacy with less residual impurities, is an area of concentrated focus for the downstream process and analytical development teams at Forge Biologics. Our teams strategically employ advanced technologies such as analytical ultracentrifugation (AUC) techniques to understand the impact of process modifications and to characterize the genetic contents packaged within an AAV capsid. Analytical ultracentrifugation analysis can identify full, empty, and partial capsids from in-process and final AAV samples, with high precision and accuracy, allowing this method to be considered the gold standard for quantifying distinct capsid populations.


Downstream Purification Strategies for Full Capsid Enrichment
CsCl Density Gradient Purification
Cesium chloride (CsCl) density gradient ultracentrifugation techniques exploit differences in buoyancies of empty and full capsids to achieve separation. CsCl and AAV move together as centrifugal force moves heavy densities of CsCl and full capsids toward the bottom of the gradient, while lighter densities of CsCl and empty capsids remain closer to the top, as seen in figure 2A. Additional species occasionally encountered are intermediate bands of partial genomes between the bands of full or empty capsids, as well as high-density bands below the full capsid band. After extended ultracentrifugation, the bands containing full capsids are collected from the tube and titered by ddPCR. The samples containing full AAV are then pooled and further purified.
Full capsid enrichment via CsCl ultracentrifugation consistently results in a final product with greater than 80-90% full capsids. In addition, this method is cost-effective and can be performed without intensive development steps, which speeds up timelines to achieve pre-clinical and early clinical milestones. However, CsCl ultracentrifugation requires open processing steps and becomes a laborious process on large production scales.
Ion Exchange Purification
An alternative method for separating empty and full particles is ion exchange chromatography (IEX) with a monolith or resin-packed column. This method differentiates AAV particles based on minute differences in the isoelectric point amongst empty, full, or partially full capsids. This results in elution from the column with the application of a conductivity gradient, as seen in figure 2B. This method allows for a fully closed system and scalable process for separating empty, full, and partial AAV particles. However, materials are costly, and the development time required to optimize the separation of vector species with a narrow range in isoelectric points can cost months.
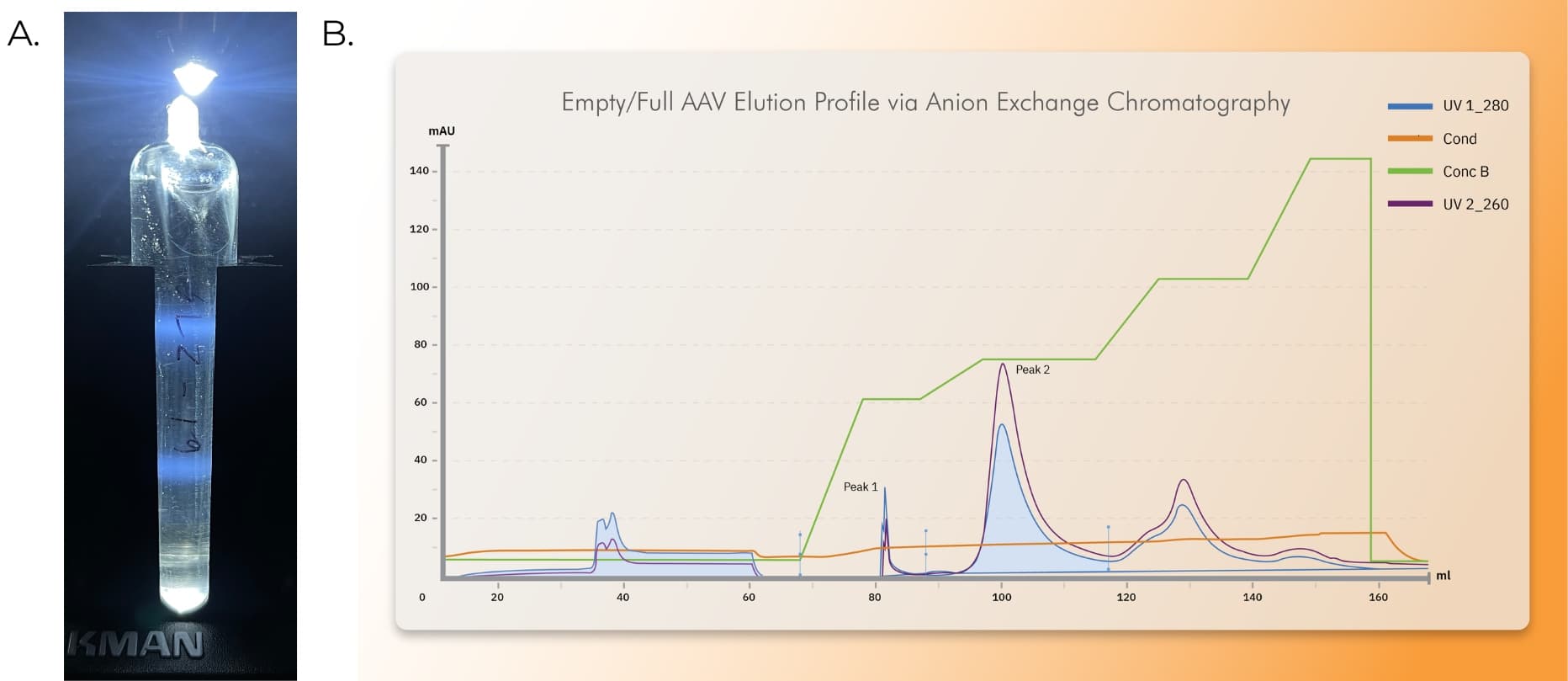
Figure 2. Empty/Full Separation via CsCl density ultracentrifugation or AEX Chromatography. AAV was produced using Ignition cells (HEK293 Suspension Cell Line) in a shake flask at 1 L working volume (A) or a 50 L bioreactor (B). Cells were triple transfected with Rep/Cap, a single stranded gene of interest under 4.7kb, and pEMBR (Ad Helper Plasmid) to produce AAV, lysed, harvested, and clarified. (A) Clarified AAV9.GFP was processed through affinity chromatography, eluted, loaded onto a CsCl gradient, and spun overnight. Visible full (bottom band) and empty banding (top band) was present after CsCl density ultracentrifugation. (B) A stylized representation of a chromatogram produced from an optimized anion exchange chromatography process. Typical preparation for an AEX run includes capture of clarified AAV vector with affinity chromatography, elution, and dilution in a buffer compatible with AEX chromatography. Vector is then loaded onto a CIMQA monolithic anion-exchange column (or alternative resin) at a concentration of ~1E+13 vg/mL and eluted over a step gradient with increasing concentrations of sodium sulfate. Peak 1 represents an enriched population of empty capsids, as indicated by UV absorbance at 280nm exceeding UV absorbance at 260nm. Peak 2 represents an enriched population of full genome containing capsids, as indicated by UV absorbance at 260nm exceeding UV absorbance 280nm.
Process and AAV Product Characterization by Analytical Ultracentrifugation
It is crucial to utilize analytical techniques that provide accurate and reproducible measurements of empty, full and/or partial capsids. Several methods that have been explored include high-performance liquid chromatography (HPLC), transmission electron microscopy (TEM), or calculation of vector genome titer by qPCR versus total capsid quantification by serotype-specific ELISA. While each method has pros and cons, limitations include long development timelines, inaccuracy, or required upfront investment into specialized equipment & expertise. One notable drawback of the commonly utilized qPCR/ELISA calculation for E/F quantification is the lack of ability to distinguish partial from full genomes leading to higher overall error. Density-based separation by AUC has proven to be a successful tool in resolving empty, full, and partial capsids and quantifying each subset with unparalleled precision and reproducibility2.
The basic principles of AUC rely on full capsids having greater buoyant density than empties, allowing them to sediment more quickly through solution during centrifugation. Based on the ratio between velocity and centrifugal field, the sedimentation coefficient (c(S)) is calculated. Full AAV capsids have a c(S) of ~100 S, empty AAV capsids have a c(S) of ~55-65 S, and partial AAV capsids have a c(S) between ~100 S and ~55 S.
To qualify the AUC methodology for each unique product, samples of AAV with varying ratios of full and empty capsids are analyzed. In a sample containing 15% empty and 85% full capsids (Figure 3A & 3B) a sigmoidal curve is seen in the sedimentation velocity (SV) scan, but the distribution plot shows distinct peaks at 55 S and 100 S, indicating the presence of both empty and full capsids, respectively. In a sample containing 30% empty and 70% full capsids, a top curve, bottom curve, and central inflection point are seen in the SV scan (Figure 3C), as well as a distribution plot with distinct full, empty, and partial species (Figure 3D). A preparation of AAV, enriched for full capsids using a CsCl ultracentrifugation method shows an SV scan with a uniform sigmoidal curve (Figure 3E), as well as a distribution curve with an elevated full peak and nearly undetectable partial or empty peaks (Figure 3F).
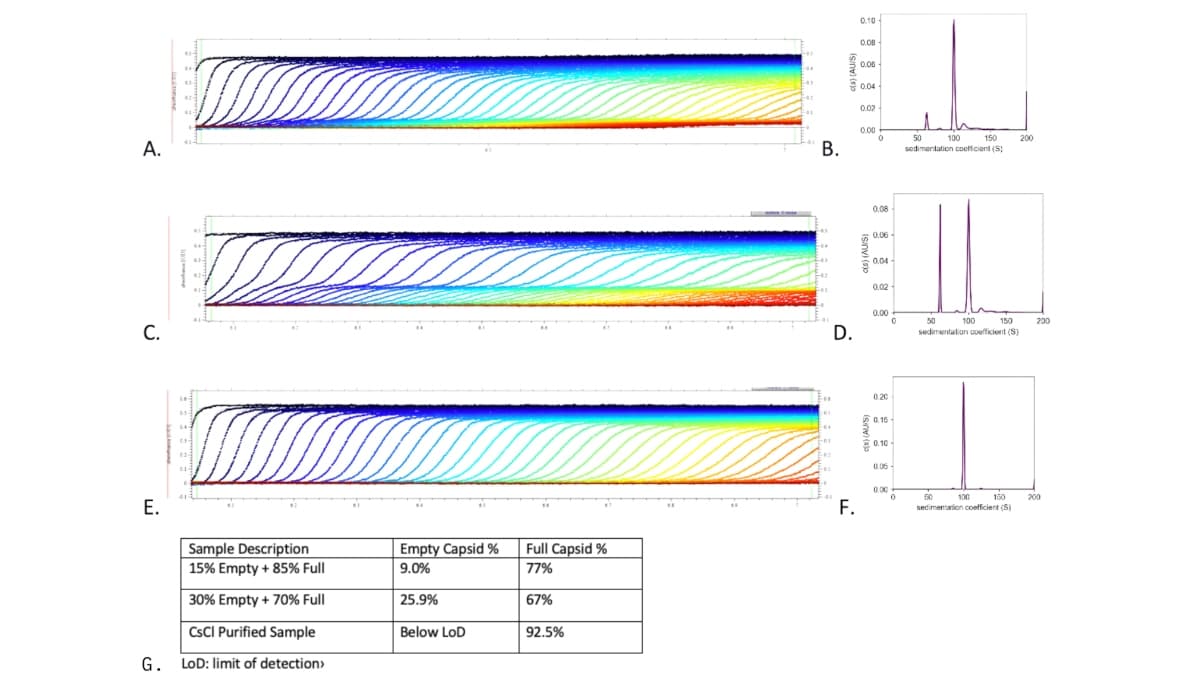
Figure 3 AUC Analysis of Mixed Full/Empty AAV or CsCl purified AAV. AAV samples in final formulation buffer were prepared for AUC analysis by diluting the sample to 0.5 O.D. in 1 mL of reference buffer. (A&B) Purified samples of empty and full AAV were mixed at 15% empty and 85% full and analyzed by AUC. (B) A stylized representation of a chromatogram produced from an optimized anion exchange (AEX) chromatography process. (C&D) Purified samples of empty and full AAV were mixed at 30% empty and 70% full and analyzed by AUC. In the SV scan, full capsids are represented at the bottom portion of the curve, partial capsids are represented in the middle inflection point of each curve, and empty capsids are seen in the top portion of the curve (C). A corresponding distribution curve shows empty capsids near 55 S and full capsids near 100 S (D). (E&F) An AAV sample purified by CsCl ultracentrifugation for the polishing step, shows a uniform sigmoidal curve on the SV scan (E). A corresponding distribution curve (F) shows a full capsid peak near 100 S with nearly undetectable levels of empty or partial capsids. (G) Sample descriptions with corresponding empty and full capsid percentages as measured by AUC and analyzed with SEDfit software.
Forge Biologics In-house AUC Capabilities
With Beckman Coulter’s Optima AUCs in-house, Forge Biologics quantifies full, partial, and empty AAV species with high precision and accuracy. By designing a systematic approach to developing downstream purification processes for each unique product, Forge achieves industry-leading separation of empty from full AAV particles. Integrated in the process development strategy, is the strategic use of AUC analysis to characterize progress and improvements beginning early in the development process. The tactical use of innovative manufacturing and analytical equipment allows Forge to efficiently produce AAV drug products of optimal purity. In addition, Forge’s attention to scalable purification methods has created readily available, platform protocols to rapidly deliver quality vector productions from less than 1 liter to 5,000 liters.
Contributing Author: Julianne Bartz, Scientist II, Forge Biologics
References:
1 Dong, B., Nakai, H., & Xiao, W. (2010). Characterization of genome integrity for oversized recombinant AAV vector. Molecular therapy, 18(1), 87-92.
2 Werle AK, Powers TW, Zobel JF, et al. Comparison of analytical techniques to quantitate the capsid content of adeno-associated viral vectors. Mol Ther Methods and Clin Dev. 2021;1(23):254-262.