
Adaptive Design Methods Offer Rapid, Seamless Transition Between Study Phases in Rare Cancer Trials
Rare cancers account for 22 percent of cancer diagnoses worldwide, yet there is no universally accepted definition for a “rare” cancer. Moreover, with the evolution of genomics and associated changes in categorizing tumors, some common cancers are now characterized into groups of rare cancers, each with a unique implication for patient management and therapy.
Adaptive designs, which allow for prospectively planned modifications to study design based on accumulating data from subjects in the trial, can be used to optimize rare oncology trials (see Figure 1). Adaptive design studies may include multiple cohorts and multiple tumor types. In addition, numerous adaptation methods may be used in a single trial and may facilitate a more rapid, seamless transition between study phases.
Figure 1. Common adaptive designs for rare oncology trials
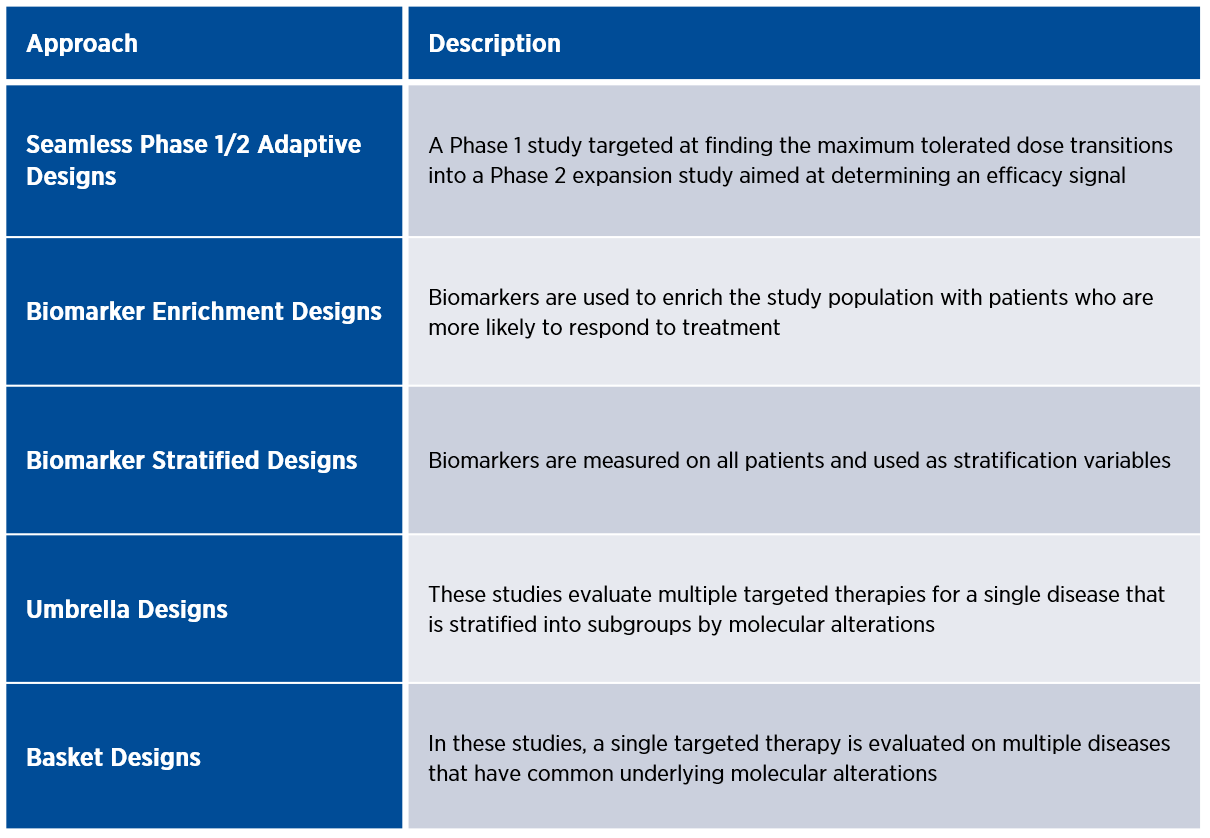
Umbrella and basket designs are examples of master protocols that utilize genomic testing to operate multiple panel studies under one overarching protocol, which increases efficiency and flexibility.
Using biomarkers in oncology clinical trials
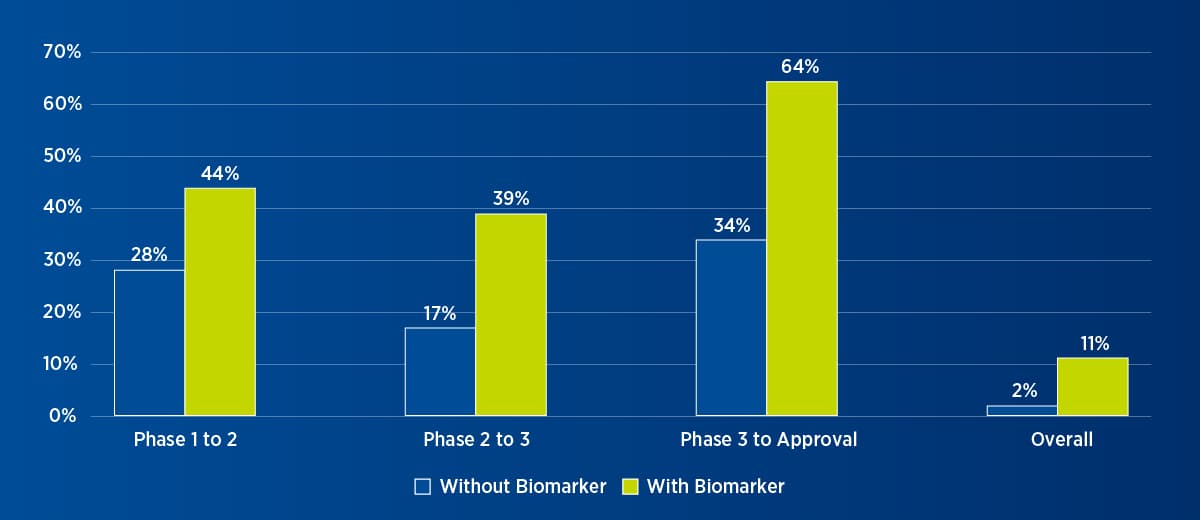
The use of biomarkers in oncology clinical trials, particularly for patient selection and monitoring of treatment response, has increased dramatically over the past five years. Significantly, research has shown that studies that use biomarkers for patient selection are not only more likely to proceed to the next stage of development but also more likely to obtain market approval (see Figure 2).
Figure 2. Oncology trial success with or without biomarker1
Biomarker testing methodologies range from simple tests that detect single mutations to complex ones that can detect a variety of gene alterations, including substitutions, deletions, duplications, and structure variance. In this universe of testing, next-generation sequencing (NGS) has been a game-changer. NGS enables the simultaneous analysis of a broad spectrum of genomic alterations. This technology can be applied to a pre-specified group of genes or gene panels, a whole exome, or even the whole genome. Coupled with dramatic decreases in the cost of sequencing and efficiencies in time- and tissue-saving, NGS analysis represents a paradigm shift that is driving both biomarker discovery and targeted drug development.
Importantly, the volume of data generated by NGS has made the interpretation of genomic data more critical than ever. Given that not all genomic information from NGS is clinically relevant, it is important to sift through the “noise” to assess the clinical significance of this data. To help in the interpretation of genomic data, some organizations have created precision medicine decision teams or molecular tumor boards. There are also increasing numbers of online genomic knowledge bases, although knowledge in rare cancer types is still disparate.
Liquid biopsies are also emerging as important biomarker tools. Liquid biopsies — which may be blood, plasma, or other body fluids — address some of the challenges associated with tissue biopsies, including availability of tissue, invasiveness, and ability to perform repeat or multiple assessments. With liquid biopsies, it is possible to perform cost-effective serial sampling, to provide dynamic and longitudinal assessment, and to perform downstream analysis of tumor markets for greater insight into the genomic landscape.
On August 7, 2020, Guardant360 received FDA approval for the first companion diagnostic to combine NGS and liquid biopsy to identify patients with specific epidermal growth factor receptor gene mutations in metastatic non-small cell lung cancer.
Empowering patient involvement
Rare diseases require strong patient advocacy and engagement. In the early 2000s, imatinib was a historic illustration of the power of the internet, and the influence patient activism can have on drug development. News of remission with imatinib treatment in a Phase 1 trial led to a patient-driven increase in clinical trial participation, which eventually led to a Phase 2 trial and subsequent approvals.
Today, technology has increased patient involvement by several orders of magnitude. Social media platforms such as Facebook and Twitter enable virtual patient knowledge-sharing on a large scale. There are Facebook groups for a majority of rare cancer types, many of which include more than 1,000 members. There are also nonprofit organizations such as Count Me In, which bring patients and researchers together to accelerate discovery and support the development of new treatment strategies.
Key takeaways
The field of oncology has changed dramatically over the last two decades. The confluence of new technology to identify rare cancers, flexible trial designs, evolving regulatory processes, and increased patient activism is advancing our understanding of cancer and accelerating the discovery of novel therapies. Premier Research offers a full spectrum of clinical research, development, and consulting services to help you optimize your rare oncology drug development. To schedule a conversation with one of our experts, click here.
1Adapted from Wong CH, Siah KW, Lo AW. Estimation of clinical trial success rates and related parameters. Biostatistics 2019;20(2):273-286.