Inventing Medicine by Accessing the Hidden Treasure in the Human Microbiome’s Dark Matter
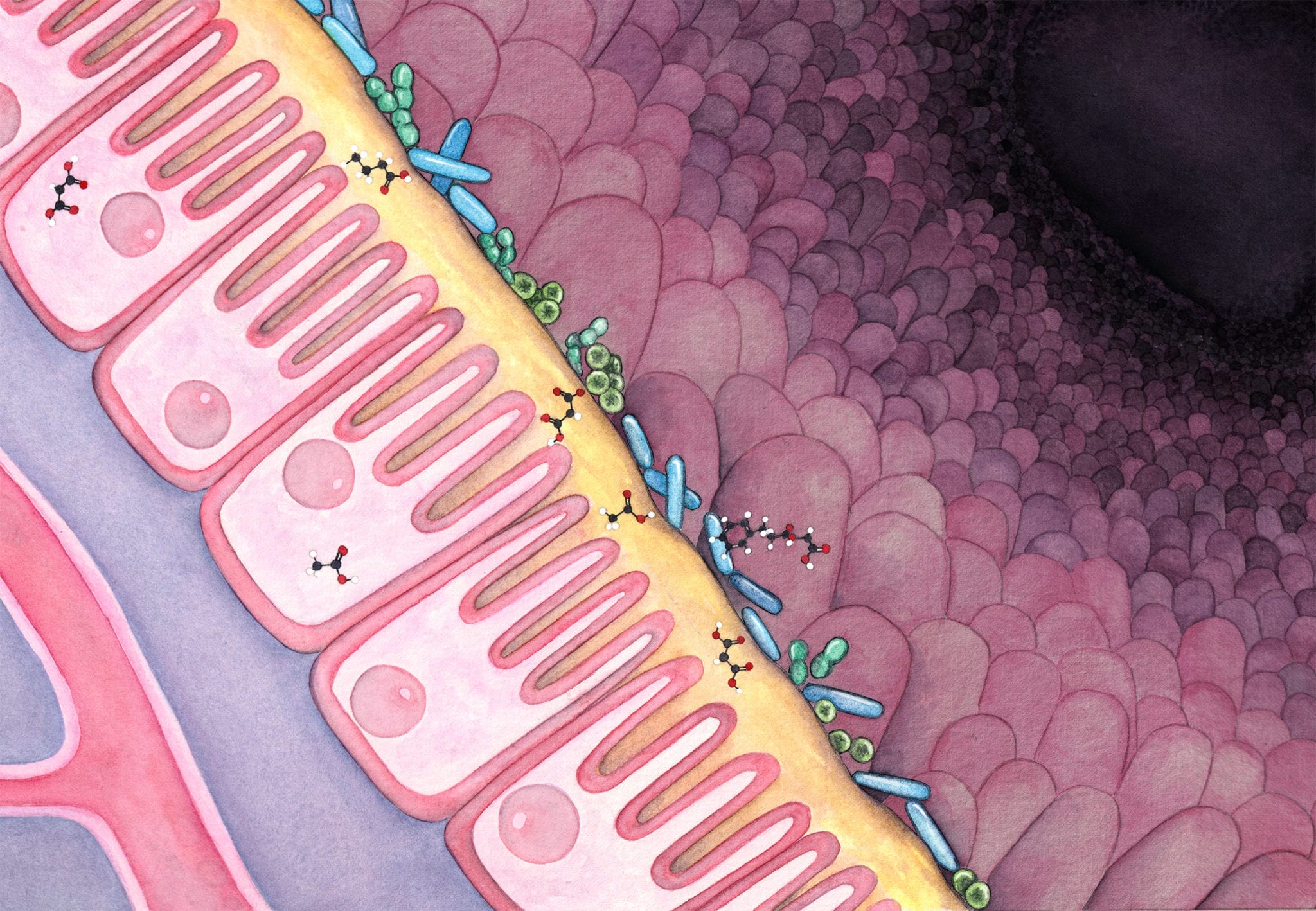
The dark matter of our microbiome
Though rapid progress has been made in understanding the microbiome’s impact on human health, a fundamental issue remains largely unsolved; and it is preventing the field from making the transition from developing therapeutics based on serendipity to inventing medicine at scale using engineering principles. This issue, at its heart, is the murky understanding of the so-called “dark matter” in the human microbiome. Collectively, the dark matter refers to the vast amount of the microbial strains that remain unculturable to researchers in the lab, and for nearly any given strain, the majority of its genome that cannot be annotated. The inaccessibility issue of the microbial strains cannot be addressed by improving our methodology of isolating and cultivating them. It is because, aside from all the logistical and biological challenges, the global diversity of the human microbiome, its rate of creating new strains, and the ease of sequencing their genomes, collectively outpace such isolating and cultivating efforts by orders of magnitude. Alongside the lack of culturability, the annotation issue of a strain genome relates to our lack of mechanistic understanding of its genomic parts, which demands vigorous molecular biology work; and traditionally, it can only be achieved after the accessibility issue is addressed.
The importance of tackling these issues cannot be overstated. The human microbiome represents the largest and the longest human trial ever: there are currently seven billion threads running globally and every single thread runs through the entire lifespan of the host. The microbes, living on the surface and inside of us, constantly respond to signals from our diet, medical treatments, mood, etc through two main interfaces: generating metabolites that exert local or systemic effects, and producing immunogenic components that elicit profound effects on our immune system. Both interfaces contain a large amount of modulators produced by the microbes. For instance, a conservative estimation of the number of secondary metabolites in the human microbiome pushed the number to be over one million; and the number of possible epitope peptides encoded by the human microbiome is even larger by at least one order of magnitude. Different from the other microbes that live in soil or marine environments, the human microbiota are comprised of members that have adapted to the human physiology as a result of long term cohabitation. The adaptations are in turn reflected in the components that operate on the above mentioned two interfaces: they are selected by evolutionary forces to be efficient at communicating or modulating their human host. All these characteristics and information are perfect for guiding drug design and drug development, yet they are concealed in the dark matter and are very difficult to access.
Accessing the opportunity space
Recent studies that probed the dark matter gave us a sense of how valuable they are. A few secondary metabolites identified were shown to be immunomodulators; a chemical screening of fecal samples have shown that one third of human GPCRs have ligand agonists produced by the microbiome; and a few microbiome-derived epitope peptides eliciting immunodominance have demonstrated therapeutic benefits in mouse models for ulcerative colitis. Moreover, on the clinical side, fecal microbiota transplantation (FMTs) and live bacteria treatments have shown that by altering the microbiome, disease states could be changed. Though delivering microbes as a new modality of medicine needs to be carefully regulated, partially for exactly the fact that they carry a substantial amount of undefined dark matter, it is without doubt that the evidence demonstrated by those procedures validated the importance of the microbiota’s components in modulating the human host. The remaining question is, how do we access this hidden treasure in a reproducible and scalable manner?
We observed that there is a big gap between our ability to survey the microbiome and our ability to engineer it; and more importantly, this gap is widening. The first principle tells us that the only way to mitigate an issue like this is to translate the surveying power into an engineering power. Fortunately, we have a tool that can facilitate this process. With rapid development in DNA synthesis, we can now transform in silico genomic information into physical form with ease. we expect this “read-write” axis to be strengthened at a fast pace, driven by the strong needs from multiple verticals such as biosynthesis of industrial chemicals, data storage by DNA, protein engineering, and genome editing by CRISPR systems. With this technology, genetic circuits embedded in the dark matter that can be conveniently and economically reproduced and tested with only in silico data. As a result of constructing this “read-write-test” axis, we can circumvent the need of cultivating strains that are very difficult to meet, and unlock the value of the vast amount of sequencing data of the microbiome. With this possibility at our horizon, I want to draw attention to the lessons we all learned from the mobile phone industry: it transformed itself from making static, hardware-driven analog phones (Nokia was the dominating player) to dynamic, feature-driven smartphones (Apple has been a key player in this wave) in jaw-dropping speed when Steve Jobs introduced iPhone to the world in 2007. The key factor here is that the introduction of the iPhone changed the cycle of introducing new features from years (making a new phone) to weeks (making a new app). Could such a transformation happen in the microbiome field?
DeepBiome Therapeutics
In the past years, most microbiome companies have been racing each other to arrive at magic strain(s) for diseases ranging from infections to central nervous system disorders. To discuss the pitfalls of such efforts is beyond this letter’s scope. However, as the iPhone did not grow out of a vacuum, we did not anticipate great success in the microbiome field would come from inventing drugs independently from the rest of pharmaceutical infrastructure by delivering blackbox-like live bacteria filled with undefined dark matter. In contrast, we see this as a perfect opportunity to engineer a “read-write-test” axis to translate such microbiome dark matter into clearly elucidated novel biological insight-based therapeutics, while working closely with tested pharmaceutical infrastructure framework. Founded by the Broad Institute and Harvard Medical School alumni scientists Chengwei Luo and Aleksandar Kostic, DeepBiome Therapeutics spent the first year at the Pagliuca Harvard Life Lab building its technology platform. With a seed round of $7.25M, lead by KTB, Baidu Ventures, and the WI Harper Group in mid-2019, DeepBiome started developing its therapeutic pipelines in the heart of Kendall Square, Cambridge. To date, DeepBiome’s platform has analyzed terabytes of microbiome data and generated drug candidates using in-depth analysis and screening methods led by deep learning and other novel algorithms. We have shown that a large percentage of these candidates, derived from microbiome insights, are bioactive with preferred pharmaceutical characteristics. Furthermore, because most of these candidates are directly derived from the healthy human microbiome, it is not surprising that they elicit very favorable toxicity profiles. Moreover, rather than collecting samples and trying to isolate strains from them, our platform was built with scalability in mind since day one to abide by the “read-write-test” mantra. With an initial focus on autoimmune diseases, we plan to use the same operations to branch into nearly any indication area in which the microbiome has been demonstrated to play a substantial role.
It is noteworthy to point out that, despite the fact that modality and scalability are in our favor, there remain several challenges to be tackled before we can fully unleash microbiome’s potential. One is the cost of DNA synthesis; though we believe it will eventually decrease to a point that we can build genetic circuits embedded in the dark matter without having to prioritize our screening effort, it is currently a speed limiting step. Another one is the screening approach itself, it is of tremendous value to engineer a unified, disease-independent platform; and we observe a few emerging technologies, such as L1000, organoids, and single cell sequencing, the could potentially offer a viable solution. Ultimately, we firmly believe that by unlocking the novel biological insights hidden in human microbiome along the “read-write-test” axis, our second genome will offer us a plethora of new therapeutics.